Tutorial
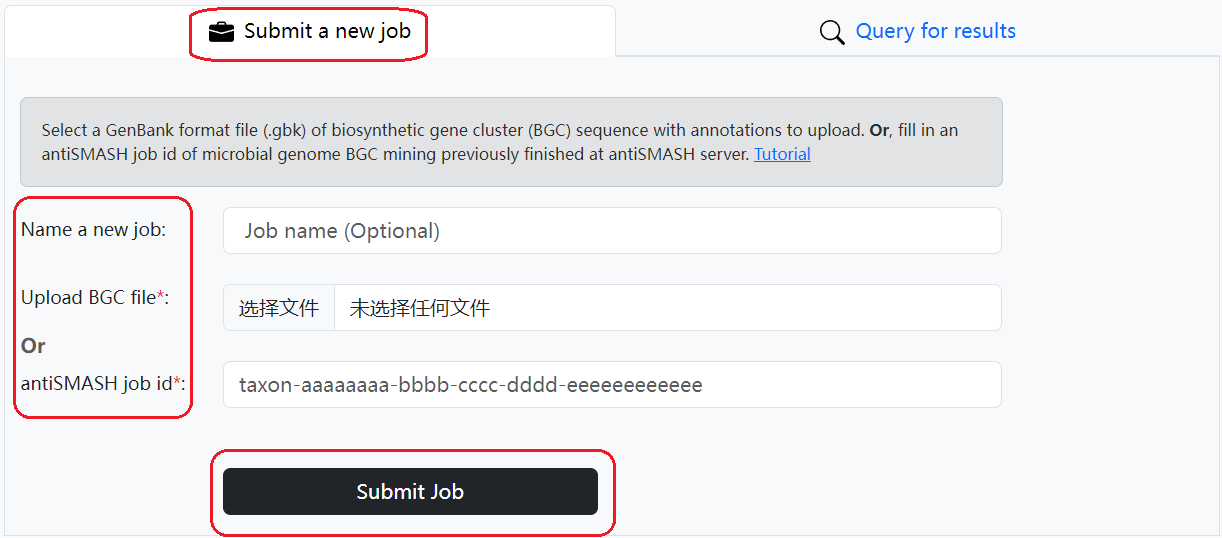
1. Submit a DeepSeMS prediction job:
Select a biosynthetic gene cluster (BGC) sequence GenBank format file and click on the 'Submit job' button to start a DeepSeMS prediction job. Or, fill in an antiSMASH job id for microbial genome BGC mining previously finished at antiSMASH server. BGC Genbank files with cluster annotation can be achieved by BGC mining tools (e.g., antiSMASH, DeepBGC, PRISM) or BGC data depositories (e.g., MIBiG). Please click on the "Load sample input" or "Download sample input" button to see the data formatting requirements. Limit maximum of 2.5 Mb file size.
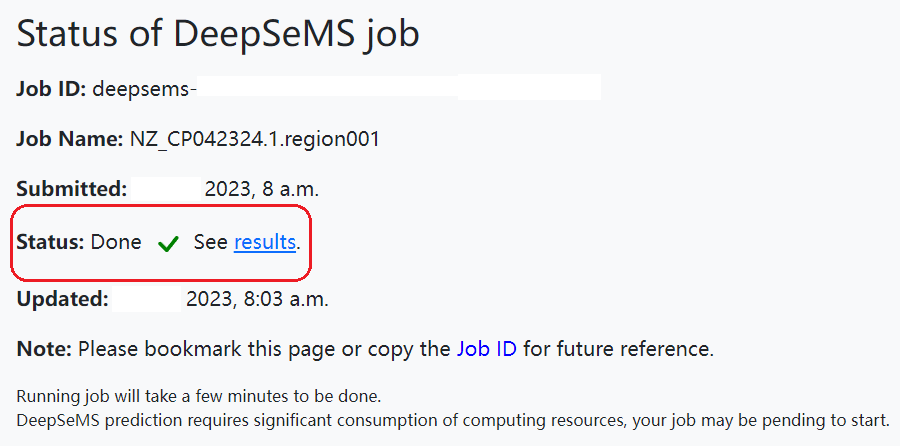
2. Query status of a DeepSeMS job:
Once a DeepSeMS prediction job was submitted successfully, the web server would automatically redirect to job status page. A job status may undergo 'Pending', 'Running' and 'Done', running job may take a few minutes to be done, it depends on the web server's load. Please bookmark the job status page or copy the 'Job ID' for future reference.
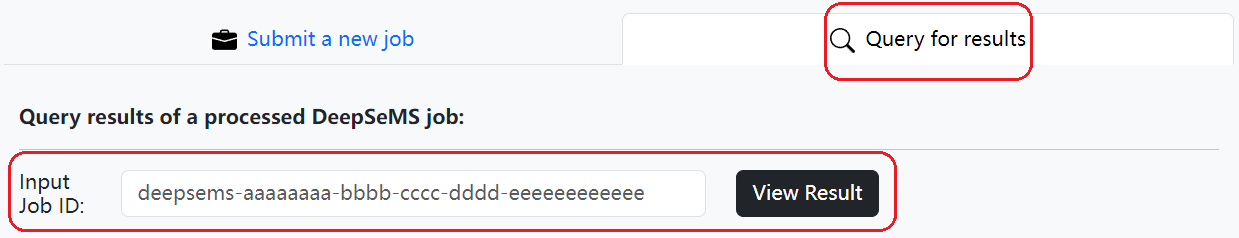
For previously submitted DeepSeMS jobs, job status or results can be queried with 'Job ID'.
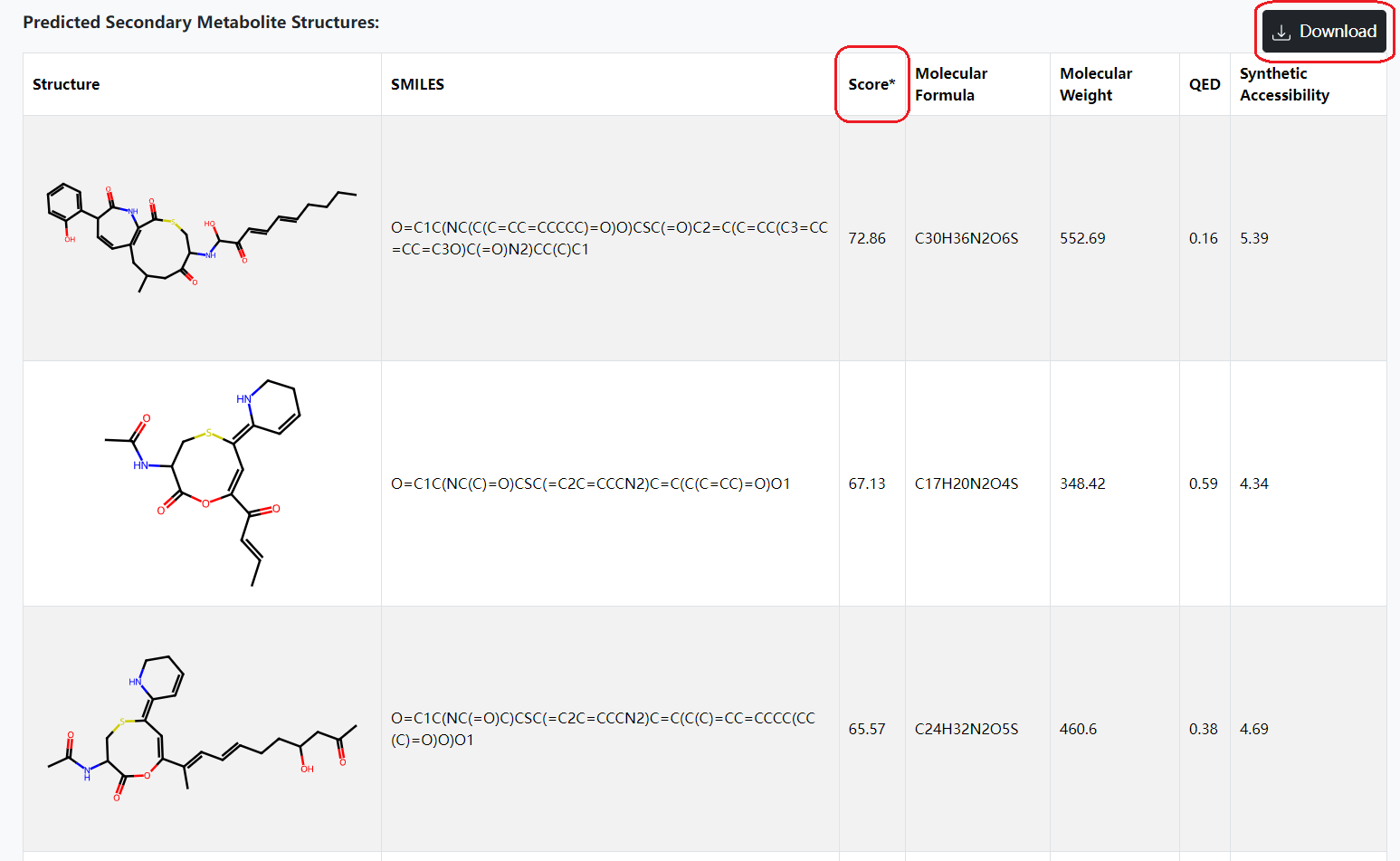
3. Understanding DeepSeMS output results:
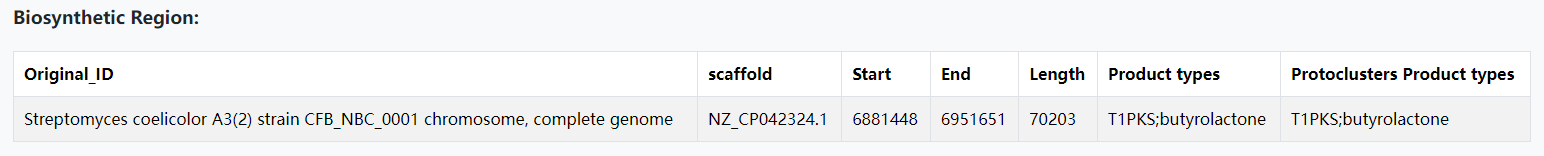
A. Please click on the "See example results" button on home page to check out the example prediction results of DeepSeMS. 'Biosynthetic Region' includes basic information about a processed BGC sequence.
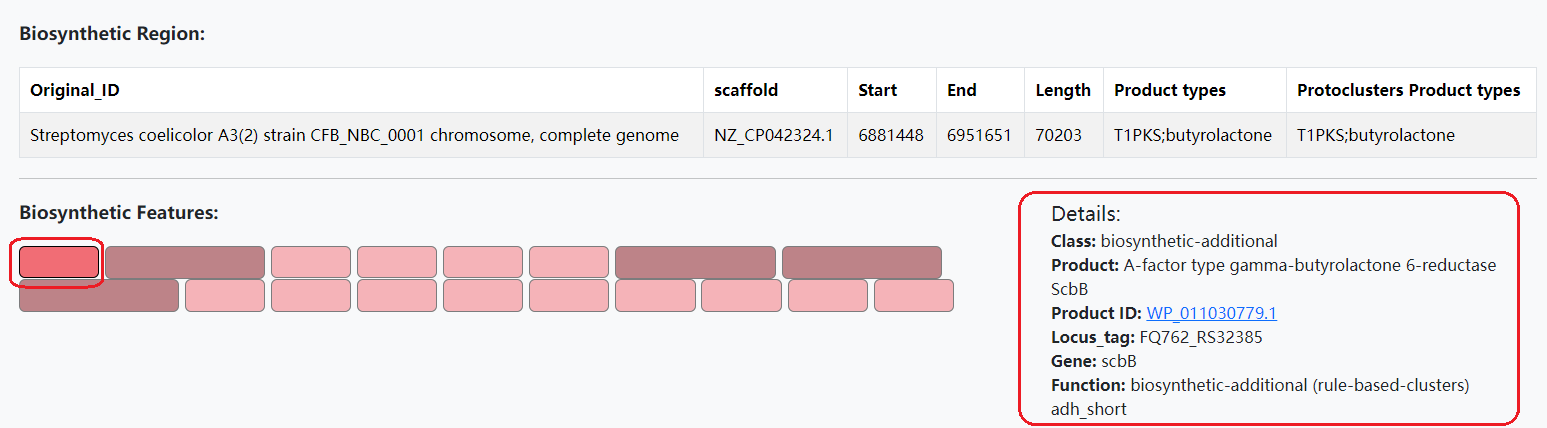
B. 'Biosynthetic Features' are biosynthetic enzymes that determine the chemical structural characteristics of secondary metabolites encoded by the processed BGC. Select a biosynthetic feature to view the details.
C. 'Predicted Secondary Metabolite Structures' that includes chemical structural predictions and property calculations of a secondary metabolite will be listed in result table, and sorted by 'Score' (Log probabilities resulted from the neural network model of DeepSeMS, only significant within each prediction). Download the result table by clicking on the 'Download' button.